Researchers at the Faculty of Engineering, University of Hong Kong (HKU), have unveiled two cutting-edge deep-learning algorithms, ClairS-TO and Clair3-RNA. These advancements promise to enhance genetic mutation detection in cancer diagnostics and RNA-focused genomic studies, marking a significant step forward in the field.
The pioneering research team, directed by Professor Ruibang Luo from the School of Computing and Data Science, has introduced these revolutionary tools aimed at transforming both clinical and research applications in genetic analysis. Utilizing long-read sequencing technologies, ClairS-TO and Clair3-RNA drastically boost the accuracy of identifying genetic mutations in complex samples, heralding new possibilities for precision medicine and genomic research. Both studies have been published in Nature Communications.
Long-read sequencing technologies capture extensive DNA and RNA sequences, offering detailed insights into genetic information. Nevertheless, navigating this data—particularly spotting mutations in challenging conditions—poses significant hurdles. The new algorithms strive to address these challenges, rendering genomic analysis quicker, more precise, and more widely available.
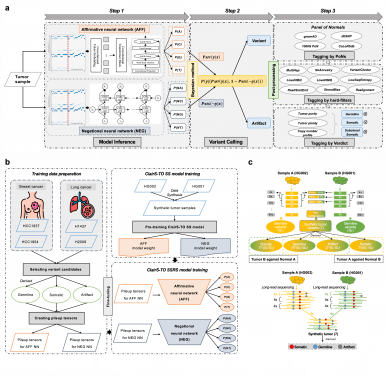
ClairS-TO confronts a vital issue in cancer diagnostics: analyzing tumor DNA without the need for matching healthy tissue samples. Traditional methods often require both types of samples for comparison, which are not always accessible. By employing a sophisticated dual-network approach—one to confirm genuine mutations and another to filter out errors—ClairS-TO removes this dependence. This breakthrough facilitates cost-effective and reliable tumor analysis, even with limited sample material, broadening access to accurate cancer diagnostics.
On the other hand, Clair3-RNA is the first deep-learning small variant caller designed specifically for long-read RNA sequencing. The challenges of RNA editing and technical sequencing errors often obscure the identification of genuine genetic variants. Clair3-RNA leverages advanced deep-learning algorithms to reliably differentiate actual mutations from background noise and editing errors, empowering researchers and clinicians to analyze gene expression and mutations with remarkable accuracy.
These algorithms expand the renowned Clair series, a suite of AI-driven genomic tools created by Professor Luo’s team. This series, which includes the widely recognized Clair3, has become a fundamental resource in computational biology. These open-source algorithms, known for their speed, accuracy, and robustness, have gained over 400,000 downloads and are extensively utilized by leading research institutes and sequencing companies worldwide, setting a benchmark for processing third-generation sequencing data.
Professor Ruibang Luo stated, “ClairS-TO and Clair3-RNA, alongside other algorithms in the Clair series, have laid a solid groundwork for deep-learning-driven genetic mutation discovery, accelerating the integration of precision medicine and clinical genomics.”
This breakthrough represents a significant advancement toward accessible, accurate, and comprehensive genetic analysis. With the potential to enhance cancer diagnosis, facilitate personalized medicine, and speed up genomic research, these innovations promise substantial benefits for both patients and scientists globally.
Link to papers:
“ClairS-TO: a deep-learning method for long-read tumor-only somatic small variant calling”
https://www.nature.com/articles/s41467-025-64547-z
“Clair3-RNA: a deep learning-based small variant caller for long-read RNA sequencing data”
https://www.nature.com/articles/s41467-025-67237-y.
About Professor Ruibang Luo
Professor Ruibang Luo serves as an Associate Professor in the School of Computing and Data Science at the University of Hong Kong. He obtained his PhD in Bioinformatics under Professor Tak-Wah Lam at HKU (2010-2015) and completed a postdoctoral fellowship with Professors Steven Salzberg and Michael Schatz at the Center for Computational Biology, Johns Hopkins University (2016-2017).
Luo specializes in bioinformatics algorithms and clinical informatics, having published more than 80 papers, ten of which have received over a thousand citations. Since 2019, he has been recognized as a Top 1% Scholar Worldwide by Clarivate Analytics, selected as one of the Top 150 Young Scholars in AI by Baidu Research, named among the Top 10 Innovators Under 35 Asia Pacific by MIT Technology Review in 2019, and included in Forbes’ 30 Under 30 Asia in Healthcare and Science in 2017.