‘An AlphaFold 4’—scientists marvel at DeepMind drug spin-off’s exclusive new AI
Isomorphic Lab’s proprietary drug-discovery model marks a significant breakthrough, yet scientists working on open-source alternatives find themselves in the dark about replicating its success.

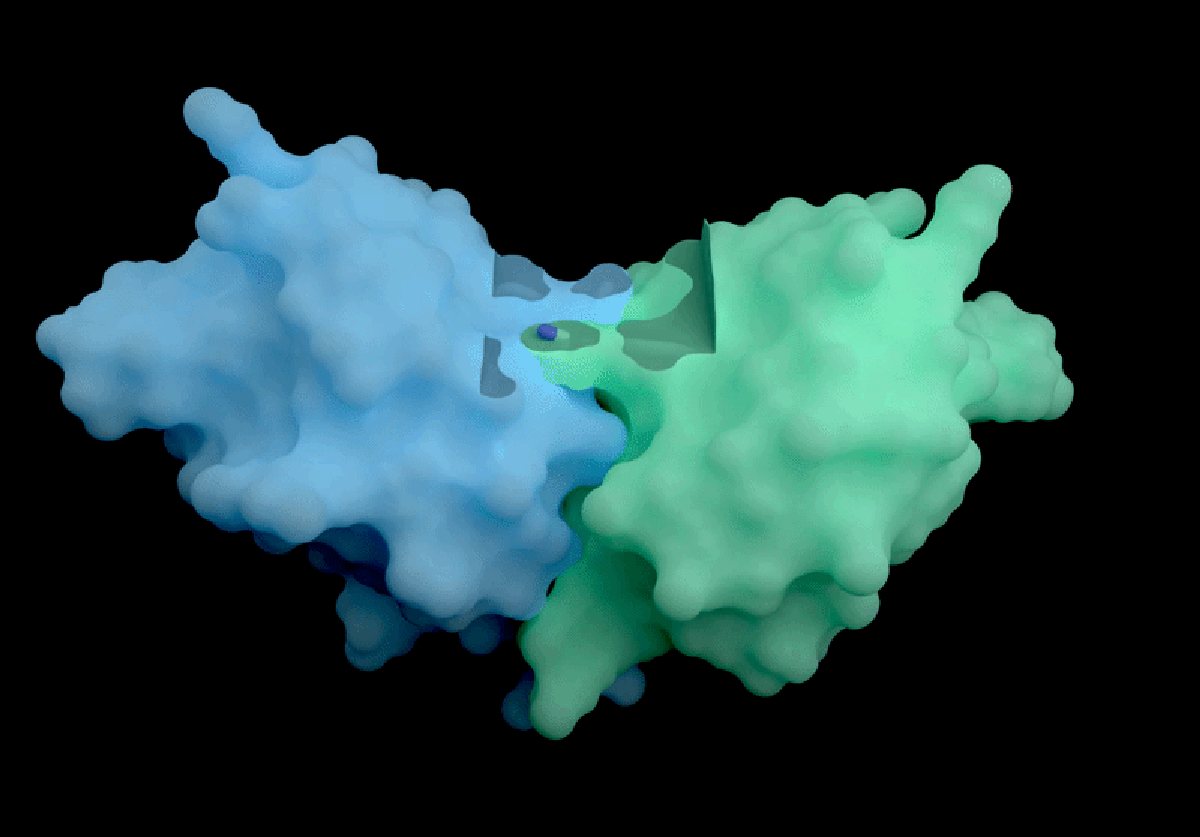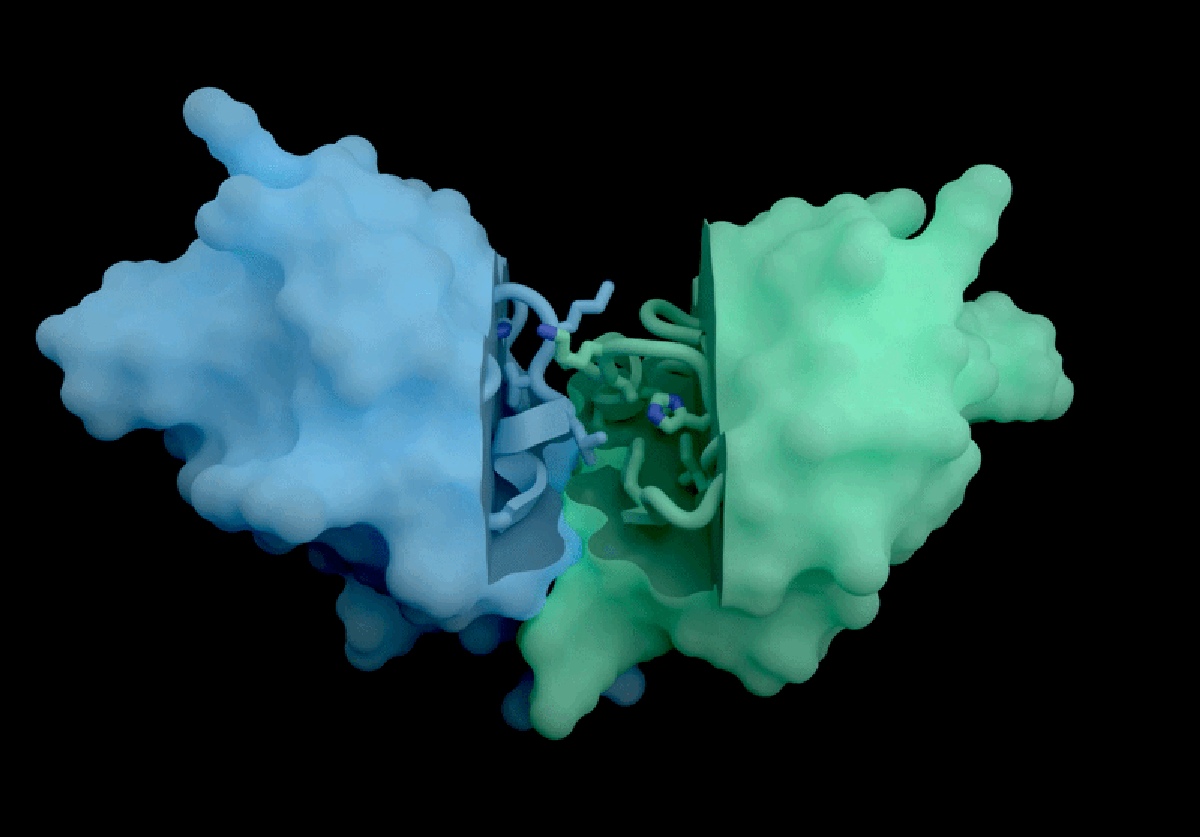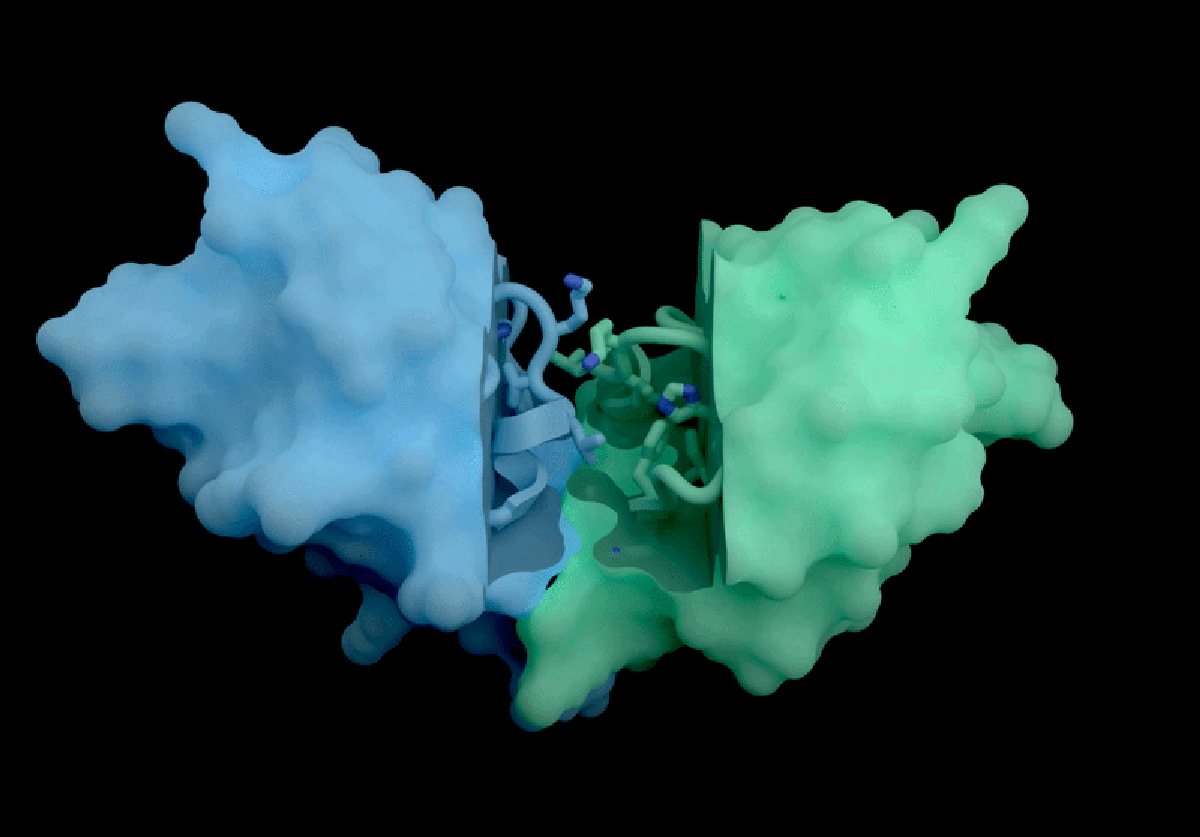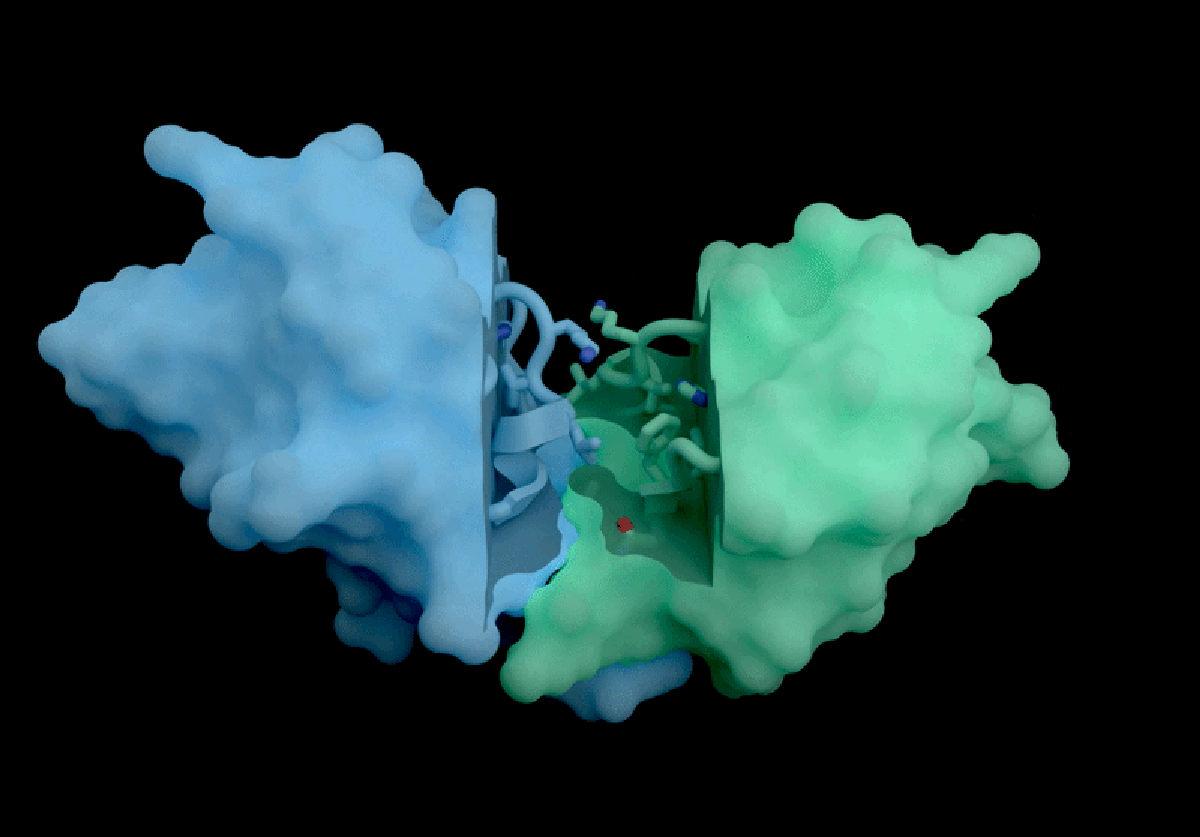
The AI tool predicts how proteins interact with potential therapeutic molecules.
Nearly two years after Google DeepMind unveiled an upgraded version of AlphaFold3 for drug discovery, its biopharmaceutical spinoff, Isomorphic Labs, has launched an even more robust AI model, which it intends to keep private.
Based in London, Isomorphic Labs recently promoted its ‘drug-discovery engine,’ named IsoDDE, in a detailed 27-page technical report released on February 10. The model has garnered attention for its accurate predictions regarding protein interactions with potential drugs and antibody structures, impressing professionals in the field.
In contrast to the AlphaFold systems, which have been made available for public research and thoroughly chronicled in scientific journals, the IsoDDE remains proprietary, providing minimal insights into its groundbreaking methodologies.
On Supporting Science Journalism
If you’re enjoying this article, consider supporting our award-winning journalism by subscribing. Your subscription fosters impactful storytelling about the discoveries and ideas shaping our world.
“It’s a significant advancement, akin to an AlphaFold4,” remarks Mohammed AlQuraishi, a computational biologist at Columbia University, who strives to create fully open-source alternatives to AlphaFold. “However, we lack critical information on the specifics.”
Drug–Protein Interactions
Developed with drug discovery as its primary focus, AlphaFold 3 differs from its Nobel Prize-winning predecessor, AlphaFold 2, by being capable of projecting the structures of proteins interacting with various molecules, including potential drugs.
Other AI models inspired by AlphaFold 3 have closely approached its performance levels while introducing new capabilities. One open-source model, Boltz-2, created by researchers at the Massachusetts Institute of Technology, accurately predicts the strength of potential drug binding to proteins, a crucial aspect in therapeutic development traditionally calculated through resource-intensive physics-based methods.
According to Isomorphic’s report, its new AI model significantly outperforms both Boltz-2 and physics-based techniques in predicting binding affinity. The AI’s predictions concerning how antibodies interact with their targets also set a new industry standard, as claimed in the report.
AlQuraishi expresses particular admiration for IsoDDE’s capability to predict drug-protein interactions for molecules that greatly diverge from the data on which the model was trained. “That represents a true challenge and indicates they must have employed some innovative approaches,” he states.
The Secret Sauce
The underlying models of IsoDDE are “profoundly different” from previous undertakings, according to Isomorphic’s president, Max Jaderberg. However, the company has no intention of disclosing the ‘secret sauce’ behind its success. “Much like other significant advancements in machine learning and AI, it comprises a blend of computational resources, data, and algorithms,” Jaderberg mentions. He hopes that the report will inspire other research teams developing drug-discovery AI.
Diego del Alamo, a computational structural biologist at Takeda Pharmaceuticals, notes that IsoDDE’s performance hints at the potential impact of industry partnerships and private structural data, although the true effect remains uncertain.
Isomorphic has secured drug-development agreements potentially valued at billions with major pharmaceutical companies, including Johnson and Johnson, Eli Lilly, and Novartis. The company also has its own internal pipeline with clinical trials on the horizon. Jaderberg asserts that various iterations of IsoDDE have been developed for use with its partners, integrating different data sources.
Michael Schaarschmidt, Isomorphic’s machine learning director, claims a “comprehensive” data strategy, which leverages publicly accessible data, synthetic training data, and data from anticipated licensing deals.
Gabriele Corso, a machine-learning scientist who co-created Boltz-2 and now leads the nonprofit Boltz in London, posits that proprietary data likely did not play a crucial role in IsoDDE’s reported success, as his team continues to uncover improvements with existing data. “This sets a new benchmark—one to aspire to surpass,” he asserts.
This article is reproduced with permission and was first published on February 19, 2026.
Support Science
If you found this article enlightening, we ask for your support. Scientific American has championed science and industry for 180 years, and this may be one of the most critical times in that history.
As a longtime subscriber, I can attest to how Scientific American has shaped my worldview. The content consistently educates and inspires, fostering an appreciation for our complex and beautiful universe. I hope it does the same for you.
By subscribing to Scientific American, you ensure our reporting focuses on meaningful research and discovery, equipping us with the resources to cover the important decisions affecting laboratories across the U.S., while also supporting both emerging and established scientists during a time when the value of science is often overlooked.
In return for your support, you will receive crucial news, engaging podcasts, insightful infographics, must-read newsletters, informative videos, challenging games, and access to the finest writing and reporting in the realm of science. You can even gift a subscription.
There has never been a more important time for us to stand up and demonstrate why science matters. We hope you will join us in this mission.